|
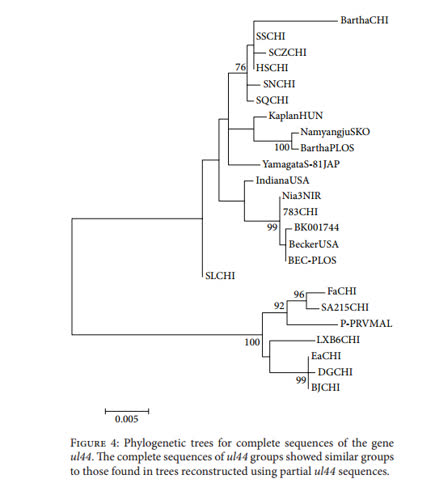
Molecular Phylogeny of Suid Herpesvirus 1 Phylogenetic analysis using only partial sequences of the
us8 gene in DNAsp identified a total of 31 haplotypes; 23 of
these haplotypes were formed exclusively by sequences from
China. Only 11 polymorphic sites were observed in Western
haplotypes (i.e., 5 singleton variable sites and 6 parsimony
informative sites). In contrast, 53 polymorphic sites were
observed in Eastern haplotypes (i.e., 38 singleton variable
sites and 15 parsimony informative sites)
|
|
Genotyping of the Pseudorabies Virus by Multiplex PCR Followed by Restriction Enzyme Analysis The BamHI-RFLP is able to discriminate the SuHV-1 into four genotypes. Genotypes I and II are distributed worldwide, and genotypes III and IV, originally described in Denmark and Thailand, respectively, were no longer reported [3]. Currently, genotype I is prevalent in populations of wild boars in Europe [8]. |
|
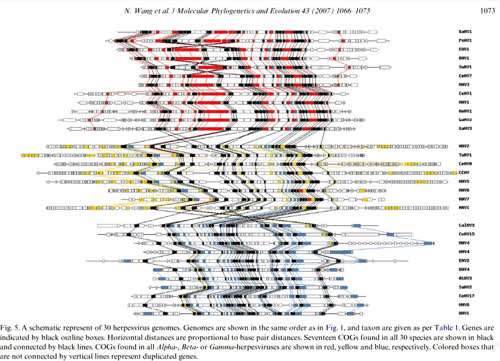
Phylogenetic analysis, genome evolution and the rate of gene gain in the Herpesviridae We used complete sequence data from 30 complete Herpesviridae genomes to investigate phylogenetic relationships and patterns of genome evolution. The approach was to identify orthologous gene clusters among taxa and to generate a genomic matrix of gene content. We identiWed 17 genes with homologs in all 30 taxa and concatenated a subset of 10 of these genes for phylogenetic inference. We also constructed phylogenetic trees on the basis of gene content data. The amino acid and gene content phylogenies were largely concordant, but the amino acid data had much higher internal support. We mapped gene gain events onto the phylogenetic tree by assuming that genes were gained only once during the evolution of herpesviruses. Thirty genes were inferred to be present in the ancestor of all herpesvirus, a number smaller than previously hypothesized. Few genes of recent origin within herpesviruses could be identiWed as originating from transfer between virus and vertebrate hosts. Inferred rates of gene gain were heterogeneous, with both taxonomic and temporal biases. Nonetheless, the average rate of gene gain was »3.5£ 10¡7 genes gained per year, which is an order of magnitude higher than the nucleotide mutation rate for these large DNA viruses |
|
Epidemiology of Aujeszky's disease The development of the disease situation in pigs in Denmark has clearly illustrated that SuHV1 has the ability to change in the degree of pathogenicity over time. Changes occurred in two steps. In the early 1960's respiratory strains developed, which were spread rapidly between herds due to animal contacts, mainly by trade, and later in the 1970's strains developed, which had an even higher degree of pathogenicity for both cattle and swine. These new strains were found to be syncytial in contrast to earlier isolates from traditional outbreaks. And that the new respiratory or syncytial strains had not been introduced from abroad was confirmed with absolute certainty by restriction fragment pattern analyses of virus DNA.
 Willy Wanker, Danish pig farmer. Name changed. File photo. |